Ozette’s data-driven panel development strategy is highlighted through a case study incorporating novel high-performance dyes into our 48-color pan-immune profiling assay.
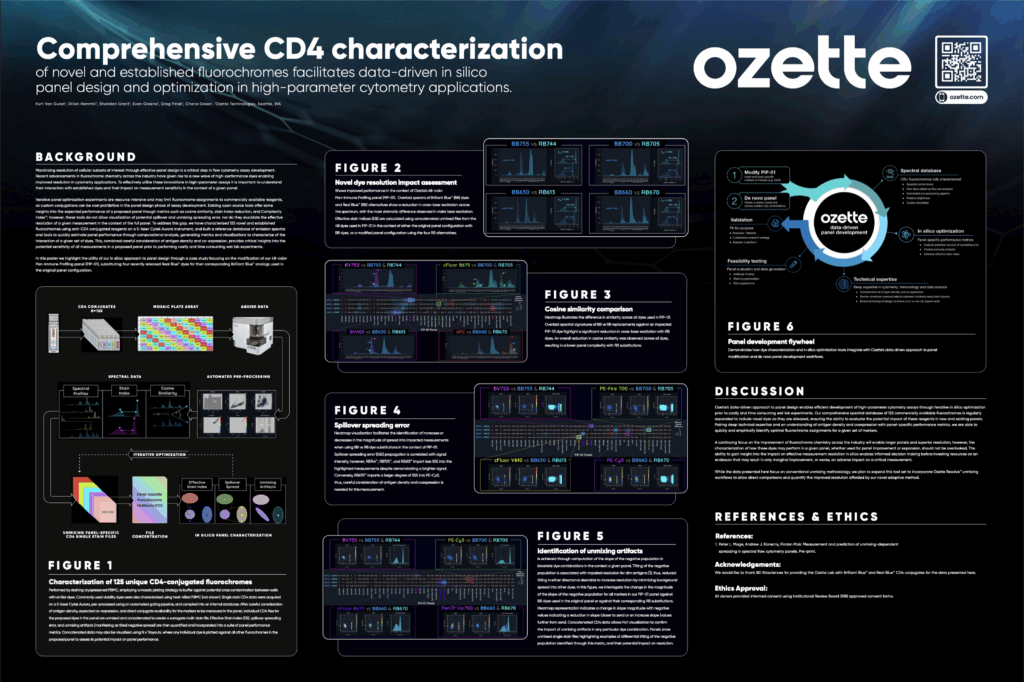
At CYTO2025, Ozette provided a detailed overview of our approach to data-driven panel design through CD4 characterization and computational performance metrics. The case study presented here demonstrates how we assessed the impact of replacing Brilliant BlueTM reagents with recently developed Real BlueTM reagents in the context of our 48-color PIP-01 assay.
Ozette DiscoveryTM elucidates the effect of blood collection matrices and cryopreservation on phenotypic subset abundance.
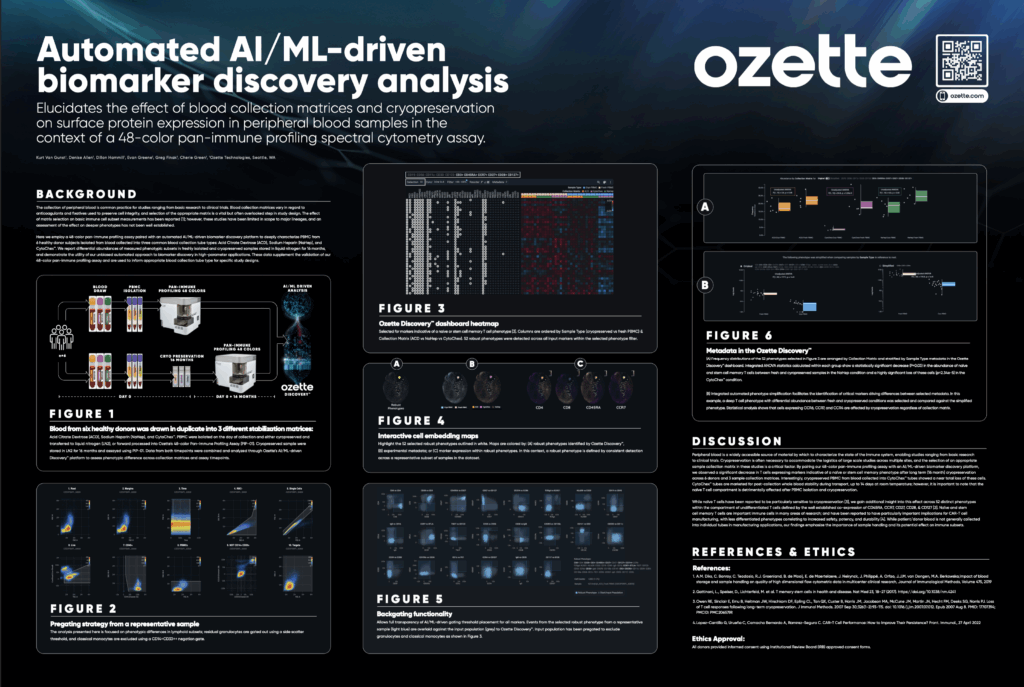
At CYTO2025, Ozette demonstrated how our ML-driven DiscoveryTM platform was used to identify differential phenotypic abundances in fresh vs. cryopreserved PBMC from blood collected into three commonly used stabilization matrices as part of the supplemental validation of Ozette’s 48-color pan-immune profiling assay.
The Insight’s in the Details: Challenges and Opportunities for BioVis Software Tools

BioVis software tools face critical challenges in handling complex analytical workflows and large-scale experimental data, particularly in enabling detailed visual exploration through computational integration, composability, scalability, and bi-directional AI/ML interactions. In this keynote, Fritz Lekschas (Head of Visualization Research) covers how modern software practices and frameworks can address these challenges, illustrated through examples from genomics, single-cell biology, and the broader BioVis community.
Aggregate Annotated Single-Cell Heatmap Visualizations

The visualization of large-scale single-cell data through heatmaps faces challenges when displaying thousands of samples and phenotypes along with their metadata, primarily due to screen space limitations and perceptual constraints. Devin Lange presents his internship work on a novel visualization technique that leverages data redundancy through intelligent aggregation of similar phenotypes and samples, enabling interactive collapsible heatmaps that can be viewed at multiple levels of detail while maintaining customizability for various use cases.
On complexity, decision making, and unintended consequences.
Scientific complexity can obscure simple errors, with potentially severe consequences. Flow cytometry, a powerful yet intricate tool, exemplifies how errors in data generation and analysis can propagate unnoticed, affecting critical outcomes in drug development and patient care. Ozette addresses these challenges by integrating validated computational methods, quality monitoring, and transparent data analysis, ensuring reliable insights that accelerate clinical research and improve decision-making.
Welcome to the Ozette Blog: Generating Quality Cytometry Data at Scale

Ozette recognizes the challenges of producing high-quality cytometry data, which stem from technical complexities and the need for diverse expertise. That is why we have launched the Ozette Blog, a platform to share our insights, expertise, and innovative solutions for generating and analyzing cytometry data at scale.
A General Framework for Comparing Embedding Visualizations Across Class-Label Hierarchies
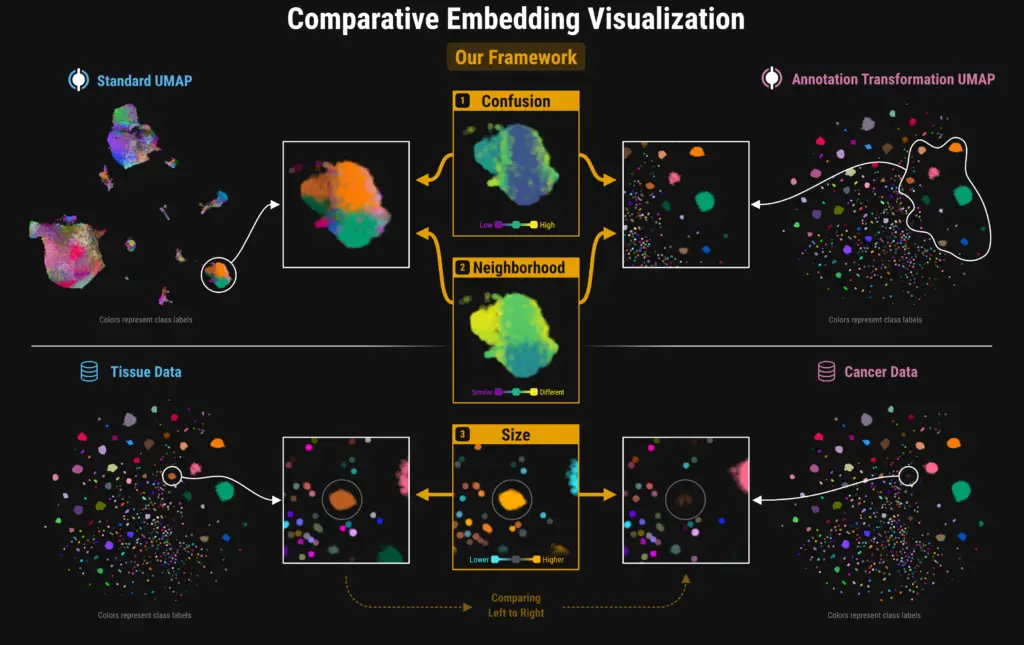
At VIS 2024, Ozette’s former intern, Trevor Manz, presented work conducted in collaboration with Nils Gehlenborg’s lab at Harvard Medical School. The presentation outlined our work on a general framework for comparing embedding visualizations across class-label hierarchies. The paper is published in the IEEE TVCG journal. Ozette uses this methodology to identify and visualize meaningful compositional differences between two single-cell datasets.
Pharmacodynamic and response biomarkers in the monotherapy arm of a phase 1 trial of CTX-471, a novel anti-CD137 agonist antibody

At SITC 2024, Ozette’s partner, Compass, presented findings from the monotherapy arm of their CTX-471 phase 1 trial. A novel anti-CD137 agonist antibody, CTX-471 treatment elicited changes consistent with increased anti-tumor immunity, including changes in natural killer cell activation status and tumor-specific expression of markers. These data identify potential biomarkers for selection of patients with a higher probability of responding to CTX-471 therapy. Read more here.
High-dimensional spectral cytometry paired with computational technology provides insights into the cellular features of healthy donor blood products to accelerate allogeneic cell therapy development
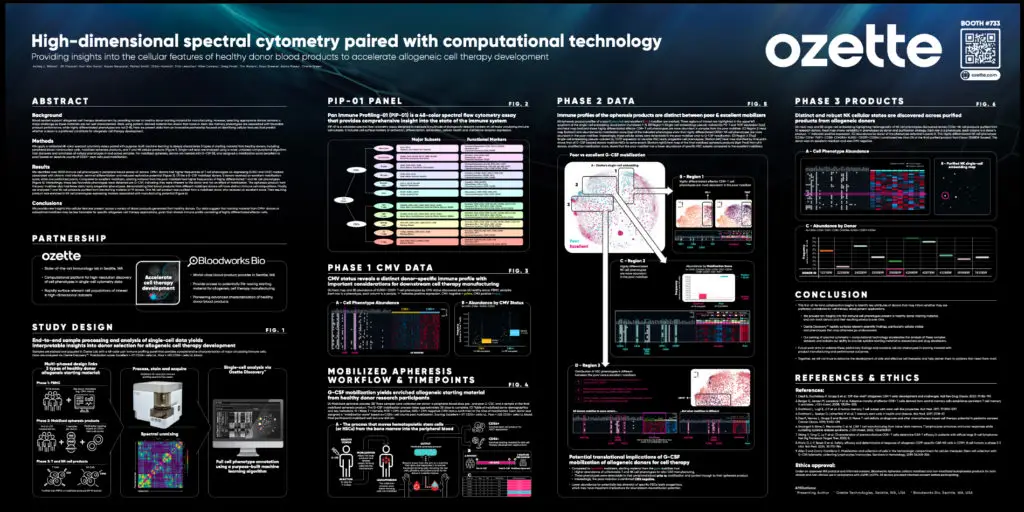
At SITC 2024, Ozette’s Head of Immunology, Ashley Wilson, presented how we use our 48-color spectral cytometry assay and AI-driven analysis platform to better identify optimal donors for allogeneic cell therapy. Analyzing immunophenotypes across different blood products from healthy donors, we found that donor characteristics, such as chronic viral infection and mobilization efficacy, can impact the suitability of blood products for allogeneic cell therapy development. Learn more by downloading the event poster here.
Pairing high-parameter spectral flow cytometry with CITE-seq using a novel automated artificial intelligence-based analysis platform to characterize immunophenotypes and cell states
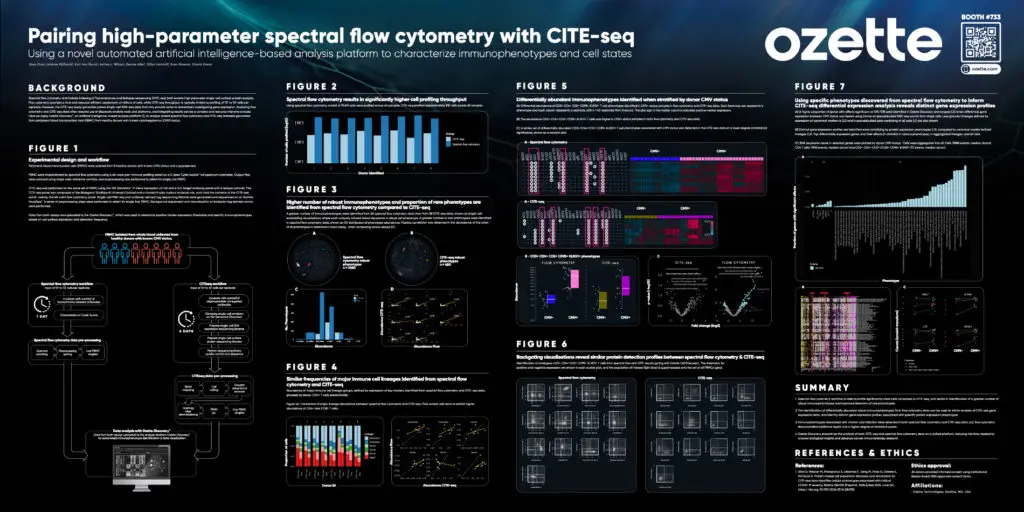
At SITC 2024, Ozette’s Zoey Zhou presented how Ozette applied our AI-driven analysis platform to streamline the analysis of paired spectral flow cytometry and CITE-seq datasets and enhance immune cell profiling. By analyzing PBMC samples from healthy donors, we identified robust immunophenotypes to inform CITE-seq differential expression analysis, revealing distinct gene expression profiles associated with defined protein expression phenotypes. Download the event poster here to learn more.